5 Import/Export
Objective:
- Learn how to import (read.csv(), read.delim()) and export tabular data (write.csv(), write.delim()) - Learn how to plot graphs using base R
We will cover:
- use data sets provided by R
- how to create a new directory/folder using dir.create()
- open several graphics windows
- save plots to file
5.1 Export Tables
** Tips for Preparing Data**
- first row = column headers (variables) and first column = row names (observations)
Row and column names
- should be unique
- shouldn’t begin with a number
- no special symbols (except underscore)
- avoid blank spaces
- avoid blank rows and comments
- use the four digit format for date: Good: 01/01/2016. Bad: 01/01/16
In Excel, save your file into .txt (tab-delimited text file) or .csv (comma separated value file) format.
Working with txt and csv files - R has built-in datasets that we may use (see datasets with data() command)
# Call data into environment with data("dataset")
# Loading DNase data
data("DNase")
# Get more info using ?
# ?DNase
# See first 10 rows with head()
head(DNase, n = 10) ## Run conc density
## 1 1 0.04882812 0.017
## 2 1 0.04882812 0.018
## 3 1 0.19531250 0.121
## 4 1 0.19531250 0.124
## 5 1 0.39062500 0.206
## 6 1 0.39062500 0.215
## 7 1 0.78125000 0.377
## 8 1 0.78125000 0.374
## 9 1 1.56250000 0.614
## 10 1 1.56250000 0.609# basic stats with summary()
summary(DNase)## Run conc density
## 10 :16 Min. : 0.04883 Min. :0.0110
## 11 :16 1st Qu.: 0.34180 1st Qu.:0.1978
## 9 :16 Median : 1.17188 Median :0.5265
## 1 :16 Mean : 3.10669 Mean :0.7192
## 4 :16 3rd Qu.: 3.90625 3rd Qu.:1.1705
## 8 :16 Max. :12.50000 Max. :2.0030
## (Other):80# unique values of a vector with unique()
unique(DNase$Run)## [1] 1 2 3 4 5 6 7 8 9 10 11
## Levels: 10 < 11 < 9 < 1 < 4 < 8 < 5 < 7 < 6 < 2 < 3R base functions for exporting/writing/saving data: * write.table() - exporting any tabular data to “.txt” file * write.csv() - comma separated values,“.csv” file
variations when comma (,) is used as decimal points instead of periods (.): write.csv2()
Arguments:
- file = absolute/local file paths with name of file
- sep = field seperators (sep argument) - character(s), controls the way splits a data table into fields/cells
- header = logical for column names
- row.names = vector of row names or number/name of column which contains the row names
# Create a Data folder
dir.create("Data")## Warning in dir.create("Data"): 'Data' already exists# Write data to txt file: tab separated values
# sep = "\t"
write.table(DNase, file = "Data/DNase.txt", sep = "\t", row.names = FALSE) #"\t" means "tab"
# Write data to csv files:
# decimal point = "." and value separators = comma (",")
write.csv(DNase, file = "Data/DNase.csv")
# Write data to csv files:
# decimal point = comma (",") and value separators = semicolon (";")
# write.csv2(DNase, file = "DNase.csv")5.2 Import Tables
R base functions for importing data
- read.table() - reading any tabular data
- read.delim() - reading tab-separated value files (.txt)
- read.csv() - comma separated value files (.csv)
variations when comma (,) is used as decimal points: read.delim2(), read.csv2()
- file = absolute/local file paths with name of file, file.choose() to choose interactively, internet address e.g. “http://www.sthda.com/upload/boxplot_format.txt”
# Read tabular data into R
# file.choose() allows you to interactively pick a data file
# df <- read.table(file.choose(), header = FALSE, sep = "\t", dec = ".")
# Read "comma separated value" files (".csv")
filename <- "Data/DNase.csv"
df <- read.csv(filename, header = TRUE)
# Read TAB delimited files
df2 <- read.delim(file = "Data/DNase.txt", header = TRUE, sep = "\t", dec = ".")
# Note to save into an object, we must assign to variable!Working with Excel files
* openxlsx package
* readxl package
Note: install libraries/packages in R using install.packages(“package_name”). Load intro environment before using with library(package_name)
Writing Data From R to Excel Files (xls|xlsx)
# # install.packages("openxlsx")
#
# library("openxlsx") # or require("openxlsx")
# # Write the first 10 rows in a new workbook
# write.xlsx(DNase[1:10,], file = "Data/DNase.xlsx", sheetName = "DNase 1", append = FALSE)
# # Add a second data set in a new worksheet with first and third column
# write.xlsx(DNase[,c(1,3)], file = "Data/DNase.xlsx", sheetName = "DNase 2", append = TRUE)Reading data From Excel Files (xls|xlsx) into R
# # Use openxlsx package
# my_data <- read.xlsx("Data/DNase.xlsx", sheetIndex = 1) Using R data format: RDATA and RDS
- Save/load variables, user-made functions and data Saving objects
- Save one object to a file: saveRDS(object, file)
- Save multiple objects to a file: save(data1, data2, file)
- Save your entire workspace (all objects): save.image() Loading objects
- readRDS(rds_file), load(RData_file)
# # Save a single object to a file
# saveRDS(DNase, "Data/DNase.rds")
# # Restore it under a different name
# my_data <- readRDS("Data/DNase.rds")
# # Save multiple objects
# save(DNase, my_data, file = "Data/data.RData")
# # To load the data again
# load("Data/data.RData")
# # Saving and restoring your entire workspace:
# # Save your workspace
# save.image(file = "Data/my_work_space.RData")
# # Load the workspace again
# load("Data/my_work_space.RData")Consider using readr package
* Reading lines from a file: read_lines()
* readr functions for writing data: write_tsv(), write_csv()
* readr for reading/writing txt|csv files: read_tsv(), read_csv(), etc
5.3 Graphing with base R
- Now that we can import our data, we can learn how to plot it.
- The purpose of base R plots is to analyze our data, not as elegant for publication purposes
plot()
- generic function, meaning it can work with different types of objects
- plot() function can be used to plot 2 variables (of equal length)
plot(x, y, type=…)
Arguments
x and y = the coordinates of points to plot
type = the type of graph to create, possible values:
“p” - points - scatter plot
“l” - lines
“b” - both points and lines
“c” - empty points joined by lines
“o” - overplotted points and lines
“s” and “S” - stair steps
“h” - histogram-like vertical lines
“n” - does not produce any points or lines
main = Title for plot
xlab = Title for x axis
ylab = Title for y axis
You can specify fonts, colors, line styles, axes, reference lines, etc. by adding graphical parameters (cex, col, lwd, etc) Read more: https://www.statmethods.net/advgraphs/parameters.html
Combine plots using par()
Read more: https://www.statmethods.net/advgraphs/layout.html
# You can plot all pairs of variables in one graph
plot(DNase)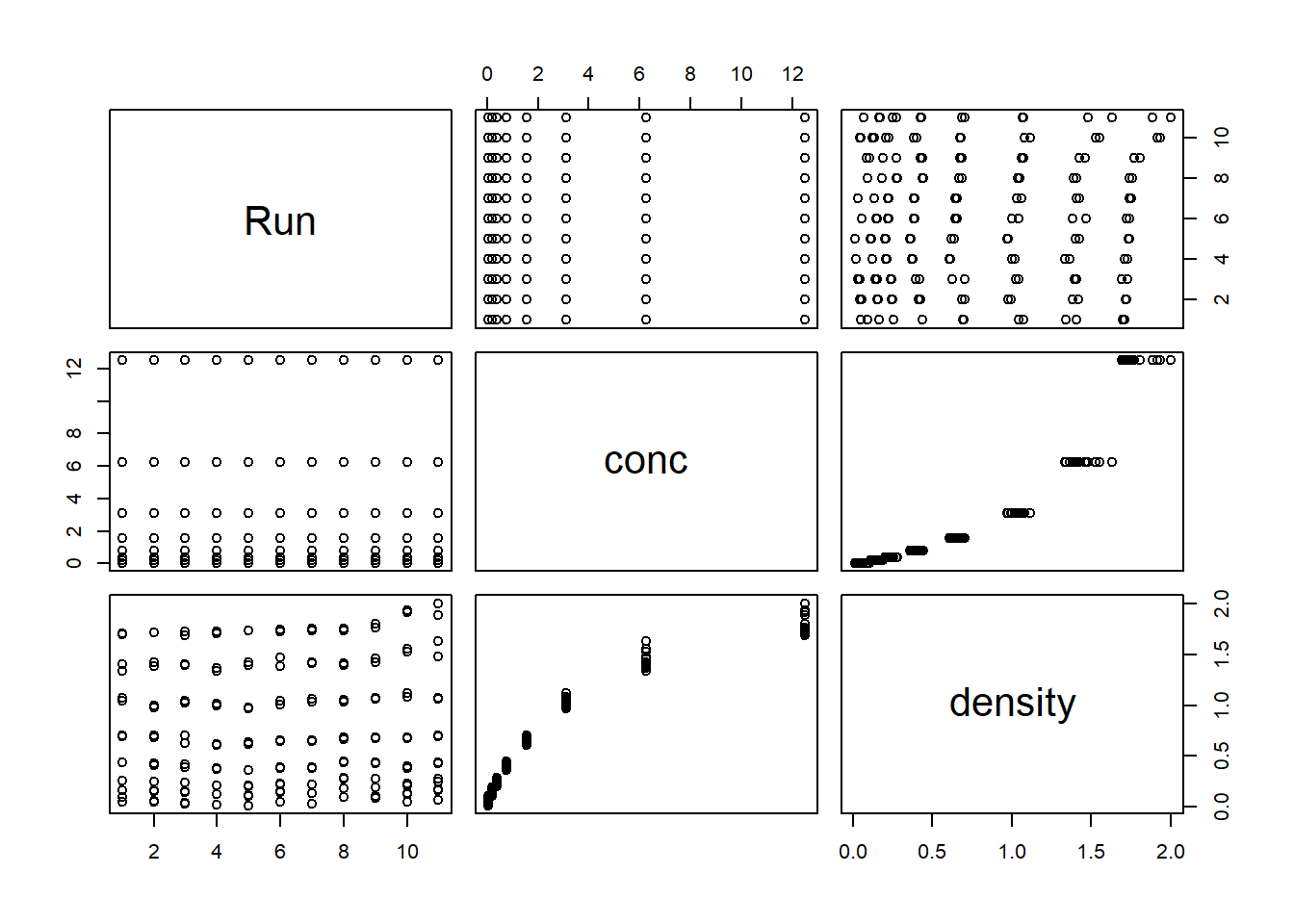
# You can also specify vectors to plot
conc <- DNase$conc; dens <- DNase$density
plot(x = conc, y = dens, type ="p") 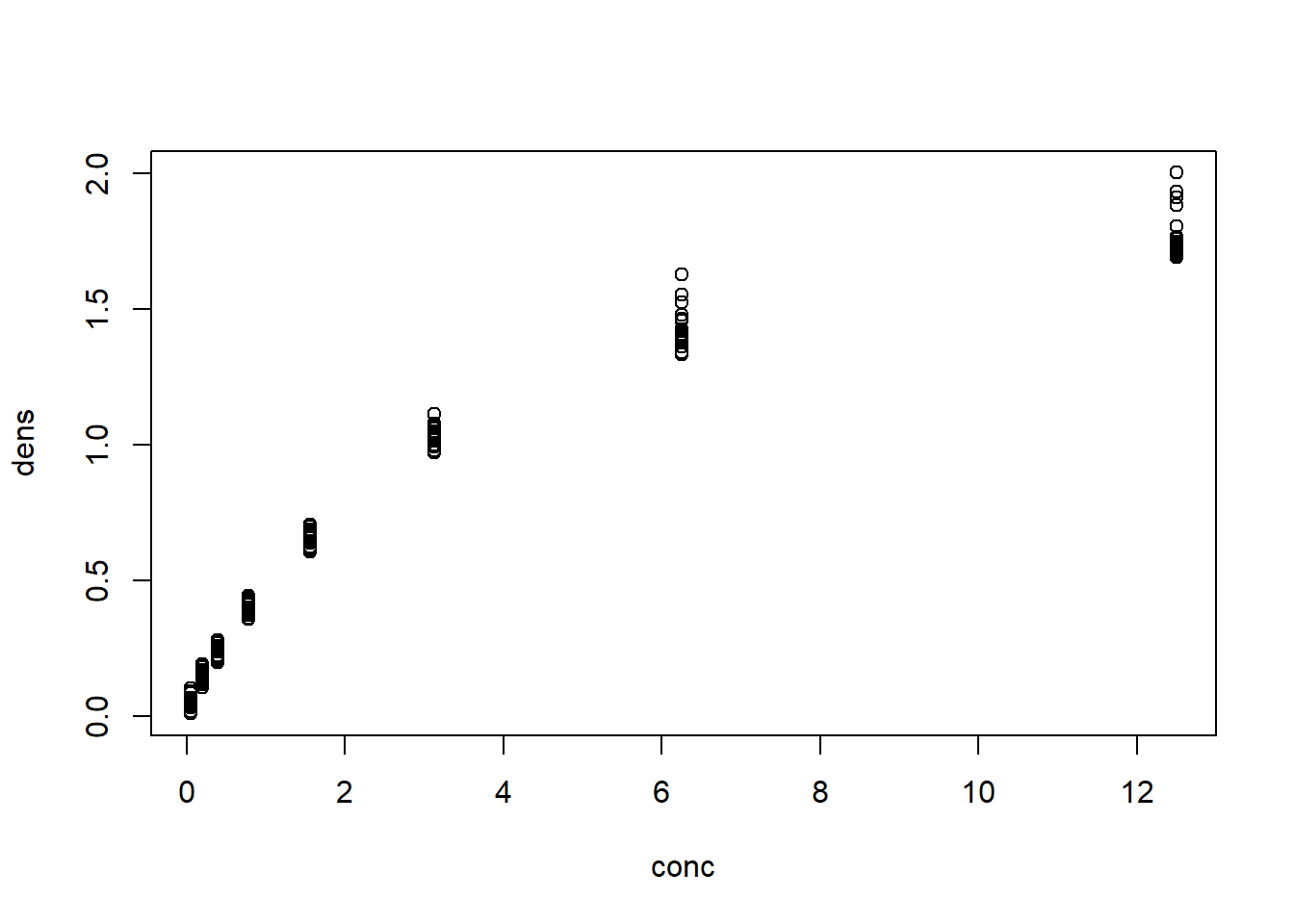
plot(x = conc, y = dens, type ="h")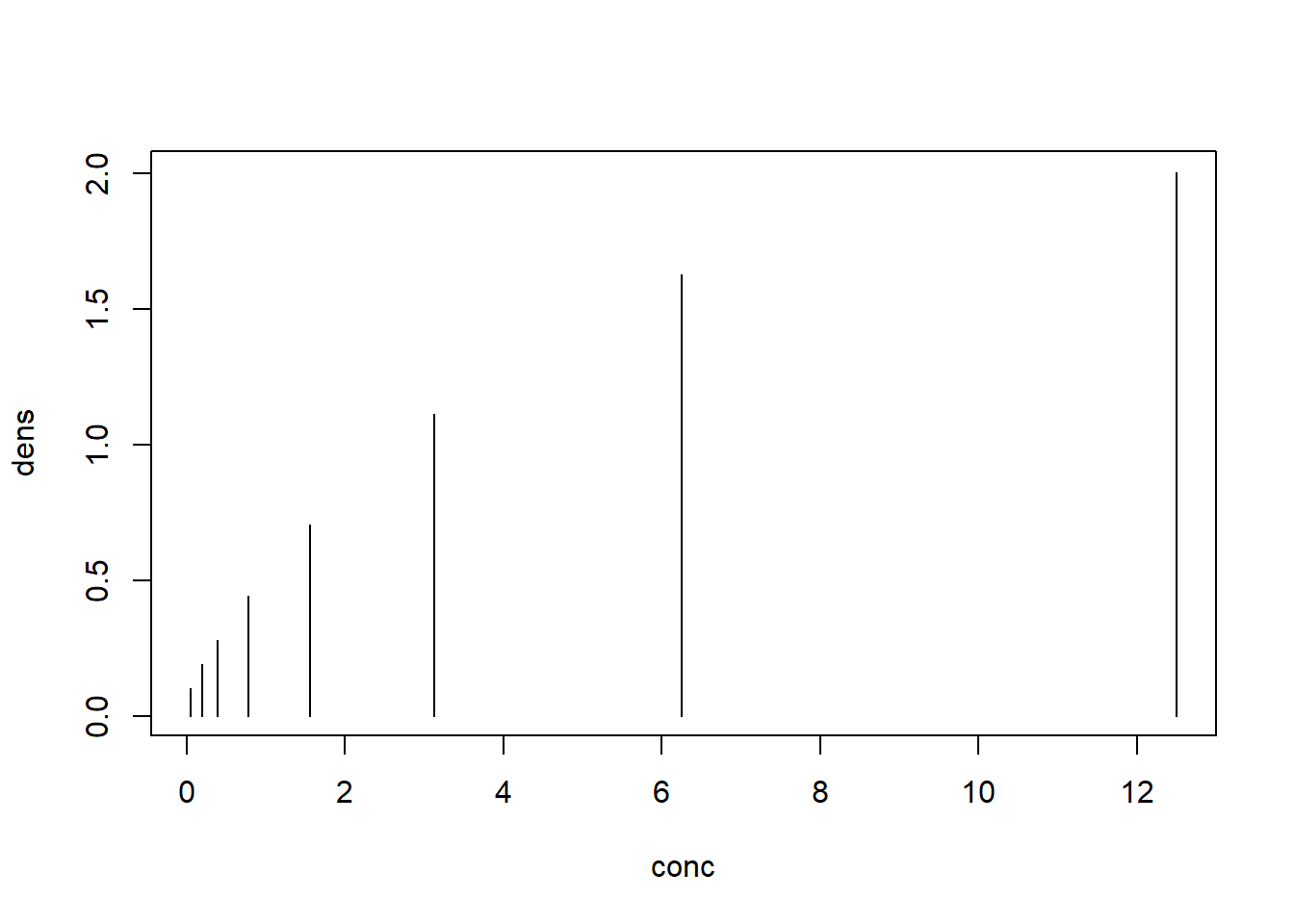
plot(x = conc, y = dens, type ="s")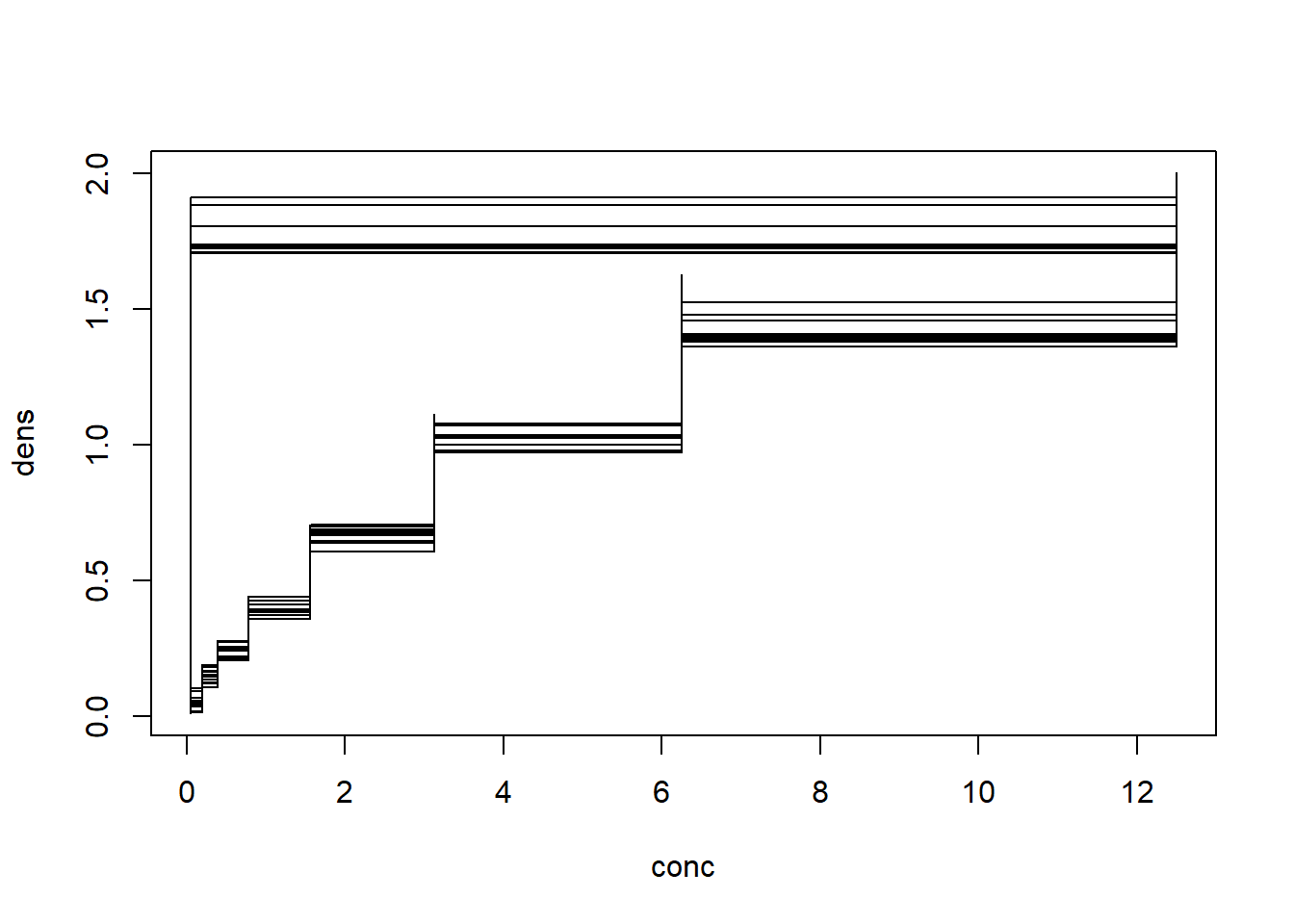
# Add title
plot(x = conc, y = dens, type ="h",
col = "red", # colour of line
lwd = 3, # width of line
xlab = "Conc", ylab = "Density", # x, y axis labels
main = "ELISA Assay of DNase") # title for plot
Tip: Open graphics windows before calling plot() to view larger plot/several graphs at once
Function - Platform In Windows, windows() or win.graph() In Unix, X11() In Mac, quartz() or x11()
# windows()
x <- seq(-pi,pi, by = 0.1)
plot(x, sin(x), main="The Sine Function")
Plot line graph with regression
Linear regression: statistical analysis technique used to determine the extent to which there is a linear relationship between a dependent variable and one or more independent variables
create simple regression using lm() or “linear model” function
2 common parameters:
+ formula: describes the model with the format “Y-var ~ X-var”, where Y-var is the dependent variable and X-var is independent variable
+ data: the variable that contains the dataset
summary() is used to find the intercept and coefficients (under “Estimate”), R^2 and p-value
Recall: y = mx + b, where m is c
Read more: https://www.r-bloggers.com/r-tutorial-series-simple-linear-regression/
# Plot
plot(DNase$conc, y <- DNase$density,
type = "p", # line graph
col = "red", # colour of line
lwd = 3, # width of line
xlab = "Concentration", # x axis label
ylab = "Density", # y axis label
main = "ELISA assay of DNase") # title of plot
# Make a linear model with the density and concentration variables
fit <- lm (density ~ conc, data = DNase)
fit##
## Call:
## lm(formula = density ~ conc, data = DNase)
##
## Coefficients:
## (Intercept) conc
## 0.2949 0.1366# Get statistics
summary(fit)##
## Call:
## lm(formula = density ~ conc, data = DNase)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.30901 -0.19640 -0.03957 0.19498 0.48056
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.29488 0.02072 14.23 <2e-16 ***
## conc 0.13657 0.00406 33.63 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.2181 on 174 degrees of freedom
## Multiple R-squared: 0.8667, Adjusted R-squared: 0.8659
## F-statistic: 1131 on 1 and 174 DF, p-value: < 2.2e-16# Add a line to the plot (new layer)
abline(fit)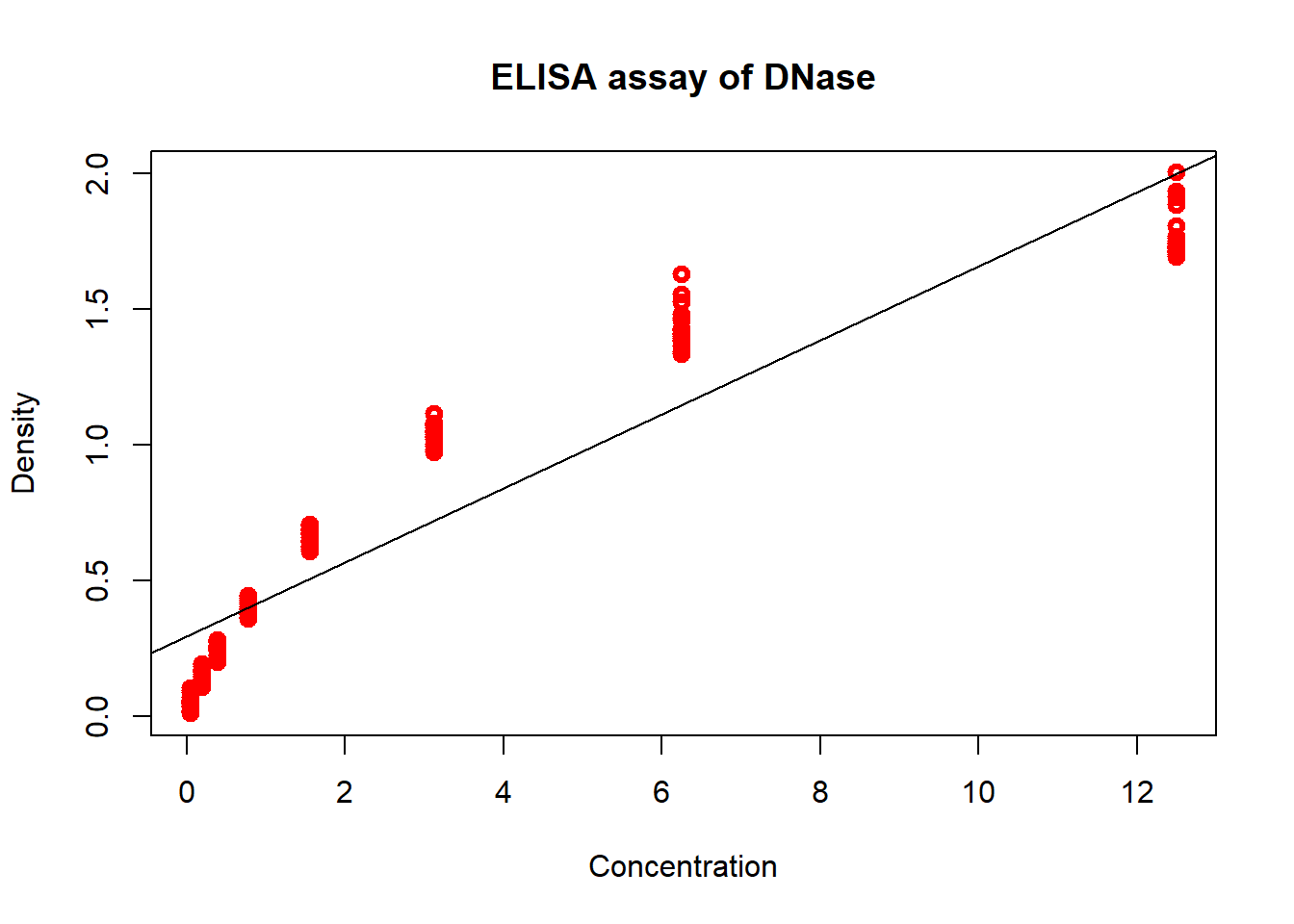
# Add text to the plot (new layer)
# text()Make a basic histogram of a numeric vector using hist()
- ref: https://rstudio-pubs-static.s3.amazonaws.com/7953_4e3efd5b9415444ca065b1167862c349.html
- Histograms: show frequencies for ranges of values
hist(DNase$density)
Saving to File
1 - Plot by exporting using “Export” in plot panel (bottom-right pane) or in window menu 2 - Plot by opening a file connection Format: type_of_file(“filename”) pdf(rplot.pdf): pdf file png(rplot.png): png file jpeg(rplot.jpg): jpeg file postscript(rplot.ps): postscript file bmp(rplot.bmp): bmp file win.metafile(rplot.wmf): windows metafile Note: Use “dev.off()” to close the connection after plotting
# Create 2 vectors with 10 elements/values
TEN_Random <- runif(10, 0.0, 1.0)
TEN_sequence <- seq(from = 1, to = 30, length.out = 10)
# Make the second value missing in y
TEN_sequence[2] <- NA
# Create a "Plots" directory in your current working directory
dir.create("Plots")## Warning in dir.create("Plots"): 'Plots' already exists# Save to png file
png("Plots/plot.png") # open "device" connection
plot(x = TEN_sequence, y = TEN_Random, xlab = "Index", ylab = "Random values") # plot
dev.off() #close connection## png
## 2Create an editable graph from R using ReporteRs package
- Editable vector graphics can be created and saved in a Microsoft document
- Read here: http://www.sthda.com/english/wiki/create-an-editable-graph-from-r-software
5.4 Practice
The “women” data set in R gives the average heights and weights for American women aged 30 to 39.
a) Print the first 15 rows to the console. (Hint: use the “n” argument in head() function)
b) Create a folder called “Data Sets” in your current working directory.
c) Write the women data frame as a csv file to the Data Sets folder (exclude row names).
d) Read this file back into R and assign it to a variable called “women.df”.
e) Plot a histogram of the heights column.
f) Find the mean and standard deviation of heights (Recall: vectors tutorial)
g) print the variables from f) in a statement “The mean and standard deviation of the heights is __ and __” (Hint: use sprintf() or paste() )
h) Plot a scatter plot, where x = height and y = weight. Relabel x and y axes to “Height (in)” and “Weight (lbs)” respectively.
i) Save f) to a jpeg file.
Solution
data("women")
# a) Print using head()
head(women, n = 15)## height weight
## 1 58 115
## 2 59 117
## 3 60 120
## 4 61 123
## 5 62 126
## 6 63 129
## 7 64 132
## 8 65 135
## 9 66 139
## 10 67 142
## 11 68 146
## 12 69 150
## 13 70 154
## 14 71 159
## 15 72 164# b) Create a folder using dir.create()
dir.create("Data Sets")## Warning in dir.create("Data Sets"): 'Data Sets' already exists# c) Write to csv using write.csv()
write.csv(x = women, file = "Data sets/women.csv", row.names = F)
# d) Read using read.csv()
women.df <- read.csv(file = "Data sets/women.csv")
# e) Plot histogram using hist()
hist(women.df$height)
# f) Find mean using mean() and standard deviation using sd()
mean.hts <- mean(women.df$height)
sd.hts <- sd(women.df$weight)
# g) Print
sprintf("The mean and standard deviation of the heights is %s and %s", mean.hts, sd.hts)## [1] "The mean and standard deviation of the heights is 65 and 15.4986942614378"paste("The mean of the heights is ", mean.hts, " and ", sd.hts, sep = "")## [1] "The mean of the heights is 65 and 15.4986942614378"# h) Plot using plot()
plot(x=women.df$height, y=women.df$weight, xlab = "Height (in)", ylab = "Weight (lbs)", main = "Avg Heights and Weights for American Women 30-39")
# i) Save as jpeg
jpeg(filename = "heights_vs_weights.jpeg")
plot(x=women.df$height, y=women.df$weight, xlab = "Height (in)", ylab = "Weight (lbs)", main = "Avg Heights and Weights for American Women 30-39")
dev.off()## png
## 2